Automated Modelling Synthesizes Brain Scan Templates
By MedImaging International staff writers
Posted on 09 Dec 2019
Automated neural network models accelerate the creation and customization of conditional atlases used in medical-image analysis.Posted on 09 Dec 2019
Developed at the Massachusetts Institute of Technology (MIT, Cambridge, MA, USA) and Cornell University (Cornell; Ithaca, NY, USA), the probabilistic model is based on one conditional neural network (CNN) that yields either universal or conditional templates, and another CNN that provides efficient alignment of the images to templates. The joint CNN is fed a random image from a dataset encoded with desired patient attributes; from that, it estimates an attribute-conditional atlas. The second CNN aligns the estimated atlas with the input image, generating a deformation field, which is then used to train a loss function to reduce deviations from a given value, thus learning to minimize differences between the learned atlas and each image.
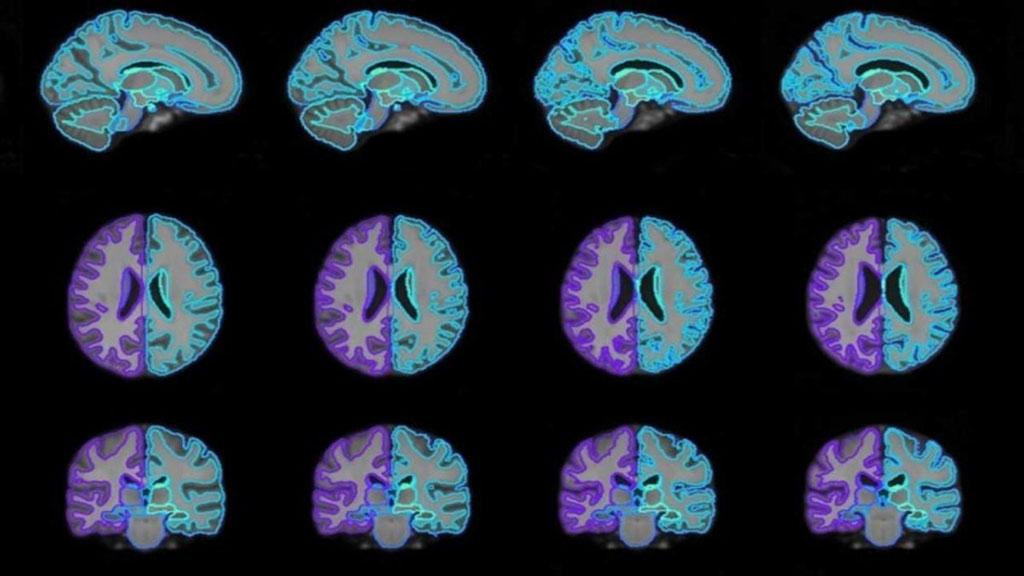
Image: On-demand brain scan templates of various ages generated using the joint CNN platform (Photo courtesy of MIT)
The networks continuously refine the atlas to smoothly align to any given image across the dataset. The result is an atlas that has learned how specific attributes, such as age, sex, and disease, correlate to structural variations across all images in a dataset. By plugging new patient attributes into the function, it leverages all learned information across the dataset to synthesize an on-demand atlas, even if that attribute data is missing or scarce in the dataset. The study was presented at the annual conference on Neural Information Processing Systems, held during December 2019 in Vancouver (Canada).
“A big dream is to build one function that can generate conditional atlases for any subpopulation, spanning birth to 90 years old. Researchers could log into a webpage, input an age, sex, diseases, and other parameters, and get an on-demand conditional atlas,” said lead author and study presenter Adrian Dalca, PhD, of MIT. “That would be wonderful, because everyone can refer to this one function as a single universal atlas reference. Atlases are central to many medical image analyses; this method can build a lot more of them, and build conditional ones as well.”
Traditional atlas-building methods run lengthy, iterative optimization processes on all images in a dataset. For example, they align all 3D brain scans to an initial (often blurry) atlas, and compute a new average image from the aligned scans. They repeat this iterative process for all images. This computes a final atlas that minimizes the extent to which all scans in the dataset must deform to match the atlas. Doing this process for patient subpopulations can be complex and imprecise if there isn't enough data available.
The atlases are available online at the MIT VoxelMorph library.
Related Links:
Massachusetts Institute of Technology
Cornell University
VoxelMorph library