Deep Learning Improves Lung Ultrasound Interpretation
Posted on 31 Jan 2024
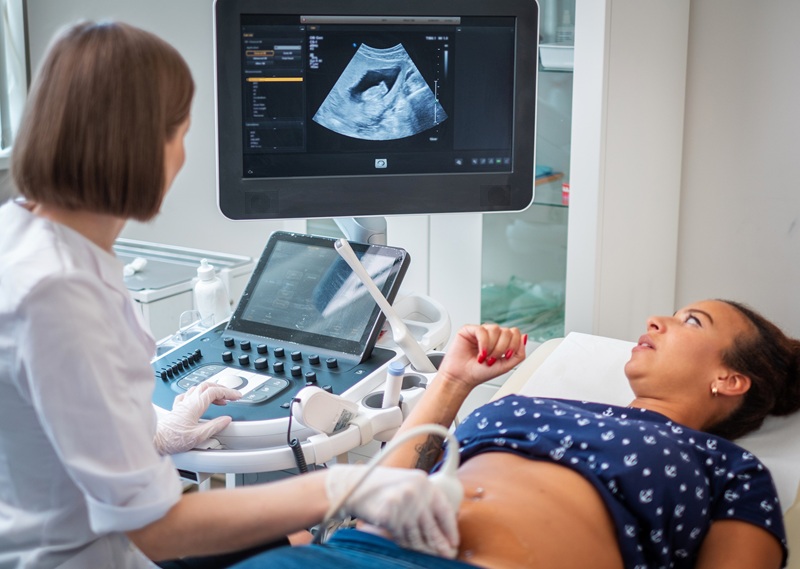
Lung ultrasound (LUS) has become a valuable tool for lung health assessment due to its safety and cost-effectiveness. Yet, the challenge in interpreting LUS images, largely due to its dependence on artefacts, leads to variability among operators and hampers its wider application. Now, a new study has found that deep learning can enhance the real-time interpretation of lung ultrasound. This study found that a deep learning model trained on lung ultrasound images was capable of segmenting and identifying artefacts in these images, as demonstrated in tests on a phantom model.
In the study, researchers at the University of Leeds (West Yorkshire, UK) employed a deep learning technique for multi-class segmentation in ultrasound images of a lung training phantom. This technique was used to distinguish various objects and artefacts, such as ribs, pleural lines, A-lines, B-lines, and B-line confluences. The team developed a modified version of the U-Net architecture for image segmentation, aiming to strike a balance between the model’s speed and accuracy. During the training phase, they implemented an ultrasound-specific augmentation pipeline to enhance the model’s ability to generalize new, unseen data such as geometric transformations and ultrasound-specific augmentations. The trained network was then applied to segment live image feeds from a cart-based point-of-care ultrasound (POCUS) system, using a convex curved-array transducer to image the training phantom and stream frames. The model, trained on a single graphics processing unit, required about 12 minutes for training with 450 ultrasound images.
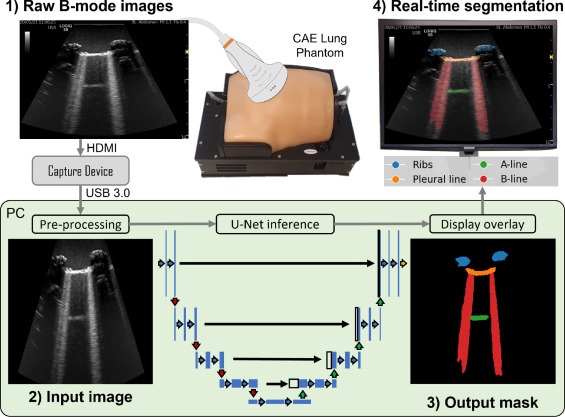
The model demonstrated a high accuracy rate of 95.7%, with moderate-to-high Dice similarity coefficient scores. Real-time application of the model at up to 33.4 frames per second significantly enhanced the visualization of lung ultrasound images. Furthermore, the team evaluated the pixel-wise correlation between manually labeled and model-predicted segmentation masks. Through a normalized confusion matrix, they noted that the model accurately predicted 86.8% of pixels labeled as ribs, 85.4% for the pleural line, and 72.2% for B-line confluence. However, it correctly predicted only 57.7% of A-line and 57.9% of B-line pixels.
Additionally, the researchers employed transfer learning with their model, using knowledge from one dataset to improve training on a related dataset. This approach yielded Dice similarity coefficients of 0.48 for simple pleural effusion, 0.32 for lung consolidation, and 0.25 for the pleural line. The findings suggest that this model could aid in lung ultrasound training and help bridge skill gaps. The researchers have also proposed a semi-quantitative measure, the B-line Artifact Score, which estimates the percentage of an intercostal space occupied by B-lines. This measure could potentially be linked to the severity of lung conditions.
“Future work should consider the translation of these methods to clinical data, considering transfer learning as a viable method to build models which can assist in the interpretation of lung ultrasound and reduce inter-operator variability associated with this subjective imaging technique,” the researchers stated.
Related Links:
University of Leeds








 Guided Devices.jpg)