Artificial Intelligence Personalizes Musculoskeletal Modeling
By MedImaging International staff writers
Posted on 11 Nov 2019
A new study shows how a deep learning tool can segment individual muscles from computerized tomography (CT) images so as to create a personalized biomechanical model.Posted on 11 Nov 2019
Developed by researchers at the Nara Institute of Science and Technology (NAIST; Japan) and Osaka University Graduate School of Medicine (Japan), the new artificial intelligence (AI) tool is based on convolutional neural networks (CNNs) that use U-Net architecture with Monte Carlo dropout, which infers an uncertainty metric, in addition to the segmentation label. By segmenting individual muscles to form a comprehensive model of the musculoskeletal system, people suffering from amyotrophic lateral sclerosis (ALS), for example, can receive a personalized rehabilitation device, and athletes can reach better performance levels.
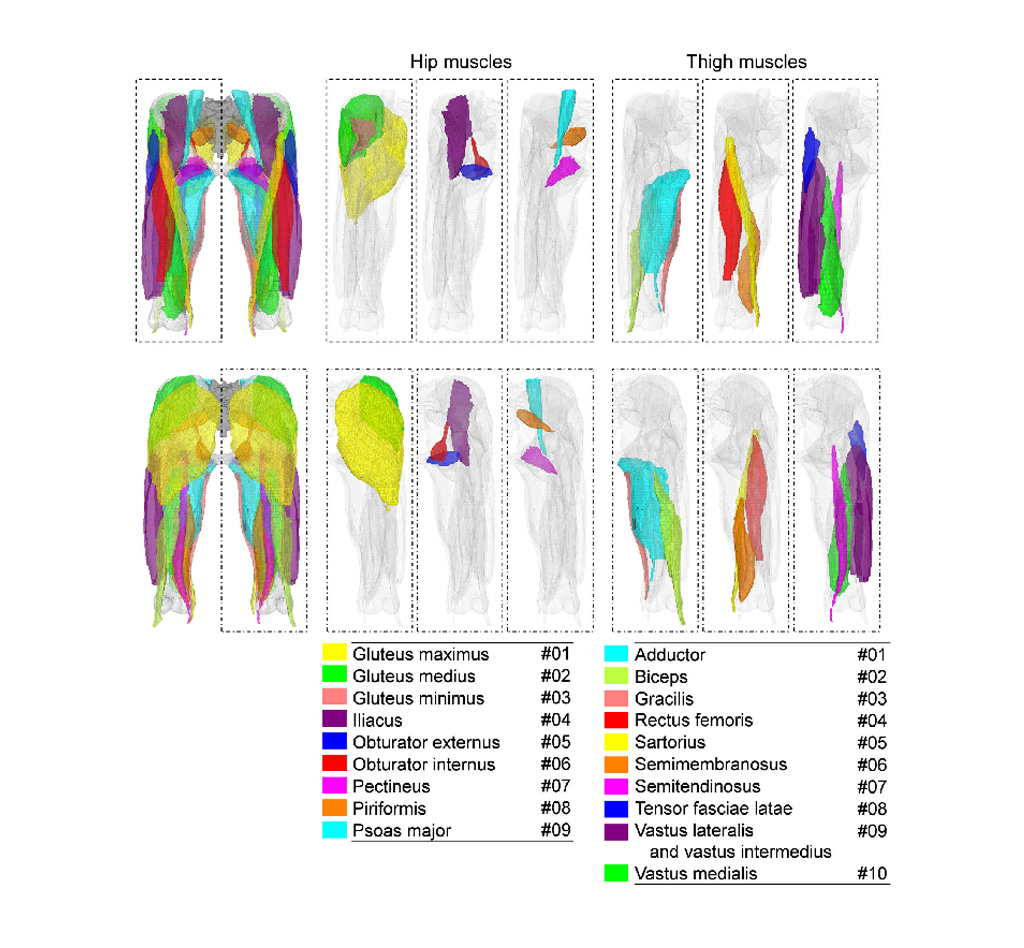
Image: Deep learning can identify individual muscles from CT (Photo courtesy of NAIST)
To evaluate the performance of the proposed method, the researchers used two data sets: 20 fully annotated CTs of the hip and thigh regions, and 18 partially annotated publicly available CTs. They found that Bayesian U-Net had better segmentation accuracy than other methods, including the hierarchical multi-atlas method, which is viewed as state-of-the-art, and did so while reducing the time to train and validate the system by a surgeon. According to the researchers, the accurate patient-specific analysis of individual muscle will aid biomechanical simulation and quantitative evaluation of muscle atrophy. The study was published on September 10, 2019, in IEEE Explore.
“Once we have the CT images, we need to segment the individual muscles for building our model. The challenge in segmenting individual muscles is the low contrast of the imaging at border regions of neighboring muscles,” said lead author Professor Yoshinobu Sato, PhD, of NAIST. “Bayesian U-Net learned the musculoskeletal anatomy to create segmentations that would have been created by experts with high fidelity and our collaborator orthopedic surgeon, Prof. Nobuhiko Sugano of Osaka University Hospital, is quite satisfied with this achievement.”
Deep learning is part of a broader family of AI machine learning methods based on learning data representations, as opposed to task specific algorithms. It involves CNN algorithms that use a cascade of many layers of nonlinear processing units for feature extraction, conversion, and transformation, with each successive layer using the output from the previous layer as input to form a hierarchical representation.
Related Links:
Nara Institute of Science and Technology
Osaka University Graduate School of Medicine